Timothy Chan Laboratory
-
Timothy Chan Laboratory
- Principal Investigator
- Research
- Our Team
- Publications
- Careers
- Research News

Timothy Chan, MD, PhD
Chair, Center for Immunotherapy & Precision Immuno-Oncology
Director, Global Center for Immunotherapy, Cleveland Clinic
Co-Director, National Center for Regenerative Medicine
Program Leader, Immune Oncology Program, Case Comprehensive Cancer Center
Sheikha Fatima bint Mubarak Endowed Chair in Immunotherapy
Location: Cleveland Clinic Main Campus
Research
The focus of the Chan laboratory is to understand the genomic basis of tumor development and treatment response. We use both large-scale genomic analyses and functional dissection to determine what drives oncogenesis. From this information, we seek to develop improved diagnostic and therapeutic modalities for human cancers.
Biography
Timothy Chan, MD, PhD, a renowned immuno-oncology and cancer genomics expert, leads the Center for Immunotherapy and Precision Immuno-Oncology, which brings together multidisciplinary experts from across the Cleveland Clinic enterprise to advance research and treatment related to the rapidly growing field of immuno-oncology.
He is an internationally recognized expert in precision immuno-oncology and a pioneer in using genomics to determine which patients will respond best to certain types of immunotherapies. He has published over 200 articles in peer-reviewed journals, made landmark discoveries in his field and received numerous awards, including the National Cancer Institute Outstanding Investigator Award in 2018.
Dr. Chan earned his MD and PhD in genetics from Johns Hopkins University, where he also completed a residency in radiation oncology and a postdoctoral fellowship in the division of tumor biology. He is board certified in radiation oncology and is an elected member of the Association of American Physicians.
Dr. Chan also collaborates with experts in the Center for Global and Emerging Pathogens Research, which is focused on broadening understanding of immunology and microbial pathogenesis with the goal of improving treatment for a variety of diseases, including virus-induced cancers.
Education & Professional Highlights
Education & Fellowships
Residency - Johns Hopkins Hospital
Radiation Oncology
Baltimore, MD USA
2007
Internship - Mercy Medical Center
Internal Medicine Internship
Baltimore, MD USA
2003
Medical Education - Johns Hopkins School of Medicine
Medicine
Baltimore, MD USA
2002
Graduate School - Johns Hopkins School of Medicine
Genetics
Baltimore, MD USA
2002
Undergraduate - Harvard University
biochemistry
Cambridge, MA USA
1995
Certifications
- Radiology - Radiation Oncology
Research
Overview
Our research group is interested in elucidating the genomic basis of tumor development and treatment response. We use large-scale genomic analyses and functional dissection to determine what drives oncogenesis. From this information, we seek to develop improved diagnostic and therapeutic modalities for human cancers.
One primary focus of our lab is to decipher how the immune system shapes tumor evolution and immunotherapy efficacy. Our group showed that tumor mutation burden helps drive anti-tumor immunity and response to immune checkpoint blockade. These findings revealed the critical role played by neoantigens in anti-tumor immunity and immunotherapy efficacy. Our work also formed the scientific foundation for the first tumor agnostic FDA approvals for cancer therapy - the use of immunotherapy in patients with mismatch repair deficient and TMB-hi tumors.
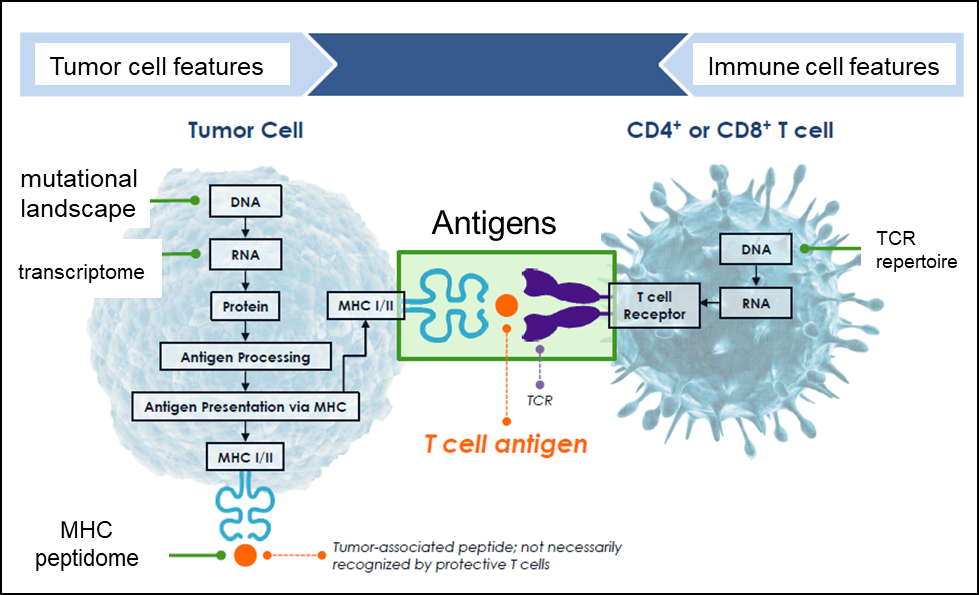
Figure 1. Our lab focuses on the dissection of tumor and immune cell processes that affect immunotherapy efficacy. We use large scale immune profiling, genomics, and functional approaches to reveal how immune cells recognize and target cancer cells.
Several major efforts are currently ongoing in the lab:
- Decoding the genetic determinants underlying response to immunotherapies
- Characterizing neoantigens and how they shape anti-tumor immunity
- Understanding how tumor heterogeneity influences immunotherapy response and how immune checkpoint therapy shapes tumor evolution
- Characterizing how cancer drivers affect the tumor microenvironment
- Development of novel immunotherapies for the treatment of cancers
We are working to understand how defects in DNA damage repair and other pathways that control genome stability lead to changes in tumor cells that are targeted by T cells. We seek to use our findings to open the door to more effective diagnostics and therapeutics for cancer patients.
Other efforts in the lab include characterizing how immune system dysregulation leads to disease processes such as COVID-induced cardiomyopathy and Parkinson’s disease.
In past years, we have identified mutations in a number of important cancer drivers and studied how these alterations lead to tumorigenesis (PARK2, FAT1, IDH1, etc). Work is ongoing to understand how mutations in these genes promote tumor development.
Seminal Contributions
Dr. Tim Chan discovered that tumor mutation burden, neoantigen burden, and DNA damage repair mutations associate with immune checkpoint blockade efficacy in patients, a discovery that supported the first tumor type agnostic FDA approvals for any cancer type. He continues to work on elucidating the molecular mechanisms of how immune checkpoint inhibitors work.
Our Team
Selected Publications
View publications for Timothy Chan, MD, PhD
(Disclaimer: This search is powered by PubMed, a service of the U.S. National Library of Medicine. PubMed is a third-party website with no affiliation with Cleveland Clinic.)
Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA*. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012 Feb 15;483(7390):479-83. doi: 10.1038/nature10866. PMID: 22343889.
Rizvi N, Hellmann MD, Snyder A, Kvistborg P, Makarov M, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, and Chan TA. Cancer Immunology. Mutational landscape determines sensitivity to programmed cell death-1 blockade in non-small cell lung cancer. Science 2015 Apr 3;348(6230):124-8. doi: 10.1126/science.aaa1348. Epub 2015 Mar 1. PMID: 25765070. PMCID: PMC4993154.
Riaz N, Havel JJ, Kendall SM, Makarov V, Walsh L, Desrichard A, Weinhold N, Chan TA. Recurrent SERPINB3 and SERPINB4 Mutations in Patients that Respond to Anti-CTLA4 Immunotherapy. Nature Genetics 2017 Feb 1;3(2):244-255. doi: 10.1001/jamaoncol.2016.1790. PMID: 27668655.
Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky J, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TA, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli E, Liu C, Harbison CT, Wang L, Ribas A, Wolchok, JD, and Chan TA*. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England Journal of Medicine 2014 Dec 4;371(23):2189-2199. PMID: 25409260.
Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi S, Martin-Algarra S, Mandal R, Sharfman WH, Bhatia S, Hwu W, Gajweski TF, Slingluff CL, Chowell D, Kendall SM, Chang H, Shah R, Kuo F, Morris LGT, Sidhom JW, Schneck JP, Horak CE, Weinhold N, and Chan TA. Tumor and Microenvironment Evolution During Immunotherapy with Nivolumab. Cell 2017 Nov 2;171(4):934-949.e16. PMID: 29033130. *Corresponding author.
Gong Y, Zack TI, Morris LGT, Lin K, Hukkelhoven E, Raheja R, Veeriah S, Meng S, Viale A, Schumacher SE, Beroukhim R, and Chan TA*. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nature Genetics 2014 Jun;46(6):588-94. doi: 10.1038/ng.2981. Epub 2014 May 4. *Corresponding author. PMID: 24793136. PMCID: PMC4251771. (featured in News and Views)
Turcan S, Makarov V, Taranda J, Fabius AWM, Wu W, Zheng Y, El-Amine N, Haddock S, Wang Y, Nanjangud G, LeKaye HC, Brennan C, Cross J, Huse JT, Kelleher NL, Osten P, Thompson CB, and Chan TA*. Mutant IDH1-Dependent Chromatin State Reprogramming, Reversibility, and Persistence. Nature Genetics 2018 Jan;50(1):62-72. PMID: 29180699.
Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein R, Makarov V, Kuo F, Kendall SM, Requena D, Riaz N, Greenbaum B, Carroll J, Garon E, Hyman DM, Zehir A, Solit D, Berger M, Zhou R, Rizvi NA, Chan TA. Patient HLA class 1 genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018 Feb 2;359(6375):582-587; PMID: 29217585.
Ganly I, Makarov V, Deraje S, Dong Y, Reznik E, Seshan V, Nanjangud G, Eng S, Bose P, Kuo F, Morris L.G.T., Landa I, Albornoz P, Riaz N, Nikiforov YE, Patel K, Umbricht C, Zeiger M, Kebebew E, Sherman E, Ghossein R, Fagin JA, and Chan TA. Integrated genomic analysis of Hürthle cell cancer reveals oncogenic drivers, recurrent mitochondrial mutations, and unique chromosomal landscapes. Cancer Cell 2018 Aug 13;34(2):256-270.e5. PMID: 30107176.. *Corresponding author.
Morris LGT, Kaufman AM, Gong Y, Ramaswami R, Walsh LA, Turcan S, Eng S, Kannan K, Zou Y, Peng L, Banuchi VE, Paty P, Zeng Z, Vakiani E, Solit D, Singh B, Ganly G, Liau L, Cloughesy TC, Mischel PS, Mellinghoff IM, Chan TA*. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nature Genetics 2013 Mar;45(3):253-61. PMID: 23354438. PMCID: PMC3729040.
Samstein RM, Lee CH Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron D, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, Motzer RJ, Hakimi AA, Voss MH, Russo P, Rosenberg J, Iyer G, Bochner BH, Bajorin DF, Al-Ahmadie HA, Chaft JE, Rudin CM, Riely GJ, Baxi S, Ho AL, Wong RJ, Pfister DG, Wolchok JD, Barker CA, Gutin PH, Brennan C, Tabar V, Mellinghoff IK, DeAngelis LA, Ariyan CE, Lee N, Tap WD, Gounder MM, D’Angelo SP, Saltz L, Stadler ZK, Scher HI, Baselga J, Razavi P, Klebanoff CA, Yaeger R, Segal NH, Ku GY, DeMatteo RP, Ladanyi M, Rizvi NA, Berger MF, Riaz N, Solit DB*, Chan TA*, Morris LGT*. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nature Genetics 2019 Feb;51(2):202-206. doi: 10.1038/s41588-018-0312-8. Epub 2019 Jan 14. PMID: 30643254. PMCID: PMC6365097. *Corresponding authors.
Mandal R, Samstein RM, Lee KW, Havel JJ, Wang H, Krishna C, Sabio EY, Makarov V, Kuo F, Blecua P, Ramaswamy AT, Durham JN, Bartlett B, Ma X, Srivastava RM, Middha S, Zehir A, Hechtman JF, Morris LGT, Weinhold N, Riaz N, Le DT, Diaz LA, and Chan TA. Genetic Diversity of Tumors with Mismatch Repair Deficiency Influences PD-1 Immunotherapy Response. Science 2019 May 3;364(6439):485-491. doi: 10.1126/science.aau0447. PMID: 31048490. PMCID: PMC6685207.
Chowell D, Krishna C, Pierini F, Makarov V, Rizvi NA, Kuo F, Morris LGT, Riaz N, Lenz TL, and Chan TA. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nature Medicine 2019 Nov;25(11):1715-1720. PMID: 31700181.
Samstein RM, Krishna C, Ma X, Pei X, Lee KW, Makarov V, Kuo F, Srivastava RM, Purohit TA, Chung J, Hoen DR, Mandal R, Setton J, Wu W, Shah R, Qeriqi B, Chang Q, Kensall S, Braunstein L, Weigelt B, Blecua P, Morris LGT, Mandelker DL, Reis-Filho JS, de Stanchina E, Powell SN, Chan TA*, Riaz N.* Mutations in homologous recombination genes BRCA1 and BRCA2 differentially affect the tumor microenvironment and response to checkpoint blockade immunotherapy. Nature Cancer 2020. http://doi.org/10.1038/s43018-020-00139-8. *Co-corresponding authors.
Zhang X, Sabio E, Krishna C, Ma X, Wang J, Jiang H, Havel JJ, Chan TA. Qa-1a modulates resistance to anti-PD-1 therapy in antigen processing defective tumors. Molecular Cancer Research 2021. PMID: 33674442.
Ho AS, Kannan K, Roy DM, Morris LGT, Ganly I, Katabi N, Ramaswami D, Walsh LA, Eng, Stephanie, Huse JT, Zhang J, Dolgalev I, Huberman K, Heguy A, Viale A, Drobnjak M, Leversha MA, Rice CE, Singh B, Iyer NG, Leemans CR, Bloemena E, Ferris RL, Seethala RR, Gross BE, Liang Y, Sinha R, Peng L, Raphael BJ, Turcan S, Gong Y, Schultz N, Kim S, Chiosea S, Shah JP, Sander C, Lee W, and Chan TA*. The Mutational Landscape of Adenoid Cystic Carcinoma. Nature Genetics 2013 Jul;45(7):791-8. PMID: 23685749.
Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan AJ, Pao W, Ladanyi M, Sander C, Heguy A, Holland EC, Paty PB, Mischel PS, Liau L, Cloughesy TF, Mellinghoff IK, Solit DB, Chan TA*. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nature Genetics 2010 Jan;42(1):77-82. PMID: 19946270.
Careers
Training at Lerner Research Institute
Our education and training programs offer hands-on experience at one of the nationʼs top hospitals. Travel, publish in high impact journals and collaborate with investigators to solve real-world biomedical research questions.
Learn MoreResearch News

Examining mutations and neoantigens in patients over their treatment with nivolumab opens the door to refining immunotherapies.

The discovery, made by a collaborative group of Cleveland Clinic researchers, opens the door to new avenues of research and reveals promising drug targets.

The study, published in Briefings in Bioinformatics, highlights how artificial intelligence can be designed to develop better immunotherapy treatments.

The honor recognizes Dr. Chan's contributions to the advances of medical science, healthcare and public health.

The gift from the United Arab Emirates will advance cancer and pathogen research globally.

Precision cancer medicine is the goal of a new multidisciplinary study, led by Dr. Tim Chan, examining radiation therapy combined with targeted therapy

Dr. Tim Chan demonstrated that pathogenic POLE/POLD1 mutations lead to improved response to immune checkpoint blockade therapy.

Dr. Chan will serve as the core investigator at Cleveland Clinic, one of four institutions working together as part of this collaborative study funded through Aligning Science Across Parkinson’s.

Researchers from the new Center for Immunotherapy and Precision Immuno-Oncology will leverage their expertise with Cleveland Clinic’s world-class resources to help advance patient care and research into new therapies related to many cancer types, including head and neck cancers.